BioNMR group of Dr. Marco Sette
Our interest is focused on the application of spectroscopic and molecular modelling techniques to solve biochemical problems. In particular, we use NMR spectroscopy either to solve biomolecular structures or to study molecular interactions.
Solution NMR provides a convenient tool for these studies since it works on systems similar to those found in cells and allows one to solve the structures of large molecules.
As an example, in my lab (in collaboration with prof. A. Battistoni, University of Rome "Tor Vergata", and prof. M. Piccioli, University of Florence) we studied the monomeric form of Salmonella superoxide dismutase (SOD), an enzyme consisting of 151 amino acids, whose structure is shown in Figure 1. Since the same enzyme is dimeric in eukaryotic systems, the structure obtained, as well as the dynamics of the backbone through 15N relaxation measurements, allows one to gain insights into the mechanism of dimerization and to suggest a hypotesis concerning its oligomerization that results in amyothropic lateral schlerosis (italian SLA).
Proteins are not isolated molecules in the cell as they interact with many other molecules, such as small ligands, ions, other proteins or nucleic acids.
We are involved in the study of protein-DNA interactions using the H-NS protein and its interaction with specific DNA fragments. This work is performed in collaboration with prof. C. Gualerzi (University of Camerino) and prof. R. Boelens and prof. A. Bonvin (both at University of Utrecht, The Netherlands). H-NS is a well known bacterial repressor but its mode of action is still matter of debate; the protein forms oligomers in solution and in vivo and it is therefore very difficult to study with state of the art biophysical techniques.
Combining molecular biology and spectroscopic approaches, we are studying the interaction of the C-terminal domain of H-NS with a specific DNA target belonging to one of the genes regulated by H-NS.
Using standard NMR approaches and a combination of labelling schemes and multidimensional filtered NMR, aimed at selecting or suppressing protein or DNA resonances, respectively, we have elucidated specific features of the binding and we are solving the structure of the protein-DNA complex (Figure 2).
A relatively new area of interest concerns the HS1.2 region of immunoglobulin. Although this region is not active in Ig production, it is conserved and several DNA regions have been shown to interact with specific proteins, thus suggesting a functional/regulative role for this region. In collaboration with prof. D. Frezza (University of Rome "Tor Vergata") and prof. A. Pastore (King's College of London, UK), we have shown that some DNA sequences belonging to the promotor regions with which these proteins interact can form an intramolecular quadruplex structure (Figure 3). Quadruplex DNA is not found only at telomeric edge of chromosomes but it is also frequently found in regulative regions of DNA. Thus, our studies suggest a functional, still unknown, role for this region. The solution structure of these structures is planned for the future.
We also apply NMR spectroscopy in biotechnological research.
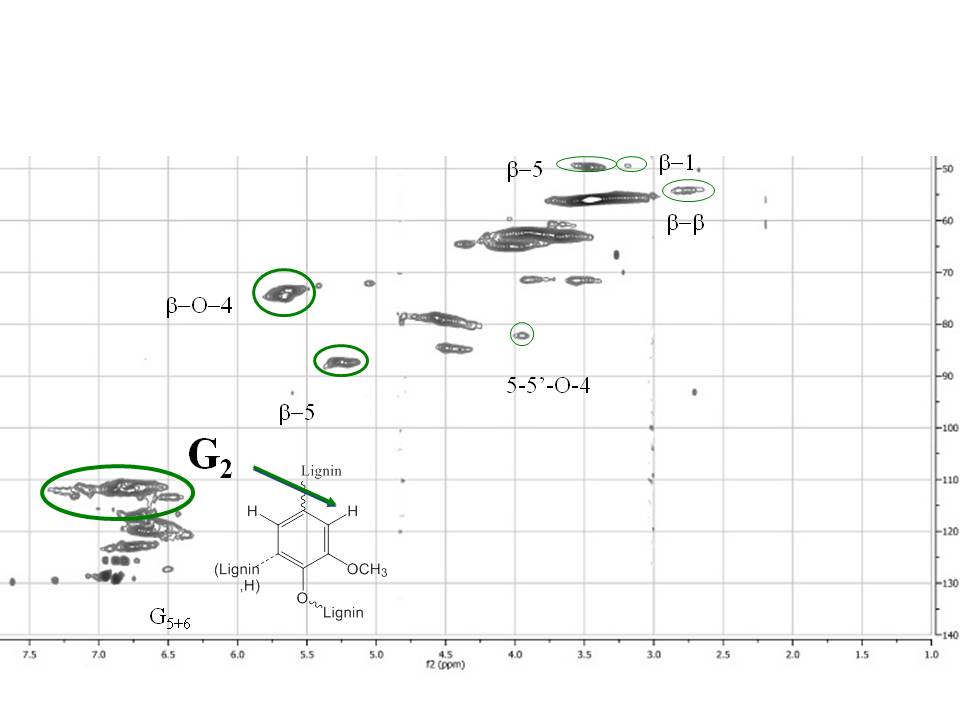
Recently (in collaboration with Dr. Claudia Crestini, University of Rome "Tor Vergata") we applied quantitative multidimensional NMR to lignin studies to unravel their structural properties. Figure 4 shows a typical quantitative 13C-HSQC collected at natural abundance. Quantitative NMR is indeed an active research area of interest in our lab (NMR day)
A future major research line (in collaboration with prof. M. Lo Bello, University of Rome "Tor Vergata", prof. M. Piccioli, University of Florence and prof. A. Bonvin, University of Utrecht, The Netherlands) concerns the interaction of glutathione-transferase with specific ligands, combining structural NMR and docking studies.
Another project (in collaboration with prof. A. Battistoni, University of Rome "Tor Vergata", prof. M. Piccioli, University of Florence and prof. A. Pastore, King's College of London, UK) concerns the zinc exchange mechanism in the bacterial system ZinT-ZnuA, a target tool for pathogenic bacteria.
Several other projects focused either on theoretical or practical applications of NMR in biological systems are being developed or under evaluation.
For our studies we use state of the art instrumentation: 600 MHz equipped with a cryoprobe, 750 MHz equipped with a multinuclear probe and 900 MHz equipped with a cryoprobe. We will also have access to a new 1200 MHz equipped with a cryoprobe (expected to be delivered in 2015).
Figure 2: A) HSQC titration of labelled C-terminal domain of H-NS in the presence of increasing amounts of specific DNA; B) observed chemical shift changes vs. sequence position; C) the C-domain of H-NS coloured according to the electrostatic potential map (negative in red and positive in blue) in which the most affected residues are indicated; D) isotope-filtered NOESY spectrum of the H-NS protein bound to its target DNA. The protein resonances have been filtered out by using an appropriate pulse sequence.
Figure 3: CD and NMR spectra of different DNA fragments showing the transition from duplex to quadruplex.
Figure 1: A) family of 30 accepted structures of reduced SodCII, displayed with MOLMOL. Structures were aligned according to best-fit superimposition of C, Ca and N atoms of residues 3-150. The metal ions are pictured in green (copper) and in grey (zinc), the b-barrel in purple, the two small b-strand in blue and the two a-helices in red. B) Backbone rmsd values for 30 conformers of SodCII. C) Superposition of the Ca trace of the mean structure of reduced SodCII (red) with X-ray structure of E.coli SOD (blue) (1eso.pdb).
Figure 4: quantitative 13C-HSQC of norway spruce milled wood lignin recorded at natural abundance.